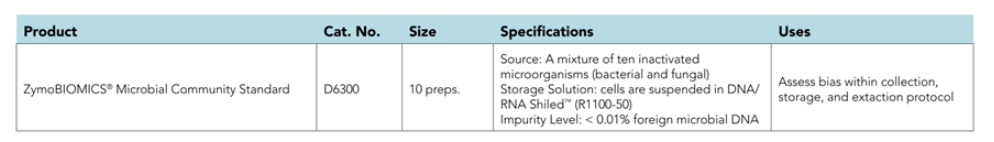
ZymoBIOMICS™ Microbial Community Standard
● Highlights
- Mock microbial community of well-defined composition.
- Ideal for the validation, optimization, and quality control of microbiomics and metagenomic workflows.
- Perfect for assessing bias of DNA extraction methods since it contains both tough and easy-to-lyse microbes.
● Description
- Microbial composition profiling techniques powered by Next-Generation sequencing are becoming routine in microbiomics and metagenomics studies. However, these analytical techniques can suffer from significant bias from collection to analysis. The ZymoBIOMICS Microbial Community Standard is designed to assess bias and errors in the extraction methods of a microbiomics workflow. The Microbial Community Standard mimics a mixed microbial community of well-defined composition, containing three easy-to-lyse Gram-negative bacteria, five tough-to-lyse Gram-positive bacteria and two tough-to-lyse yeasts. Acting as a defined input from the beginning, the Microbial Community Standard can guide construction and optimization
▶ Accurate Characterization
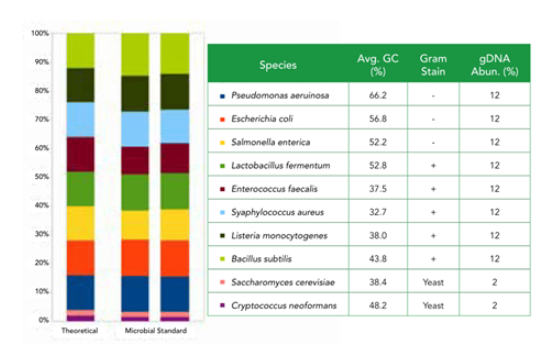
Containing three easy-to-lyse Gram-negative bacteria, five tough-to-lyse Gram-positive bacteria, and two tough-to-lyse yeasts, the ZymoBIOMICS® Microbial Community Standard is perfect for assessing bias in various DNA extraction methods. The microbial standards are accurately characterized, with a wide GC range (15%-85%) and contain negligible impurities (<0.01%), enabling easy exposure of artifacts, errors, and bias in microbiomics or metagenomic workflows.
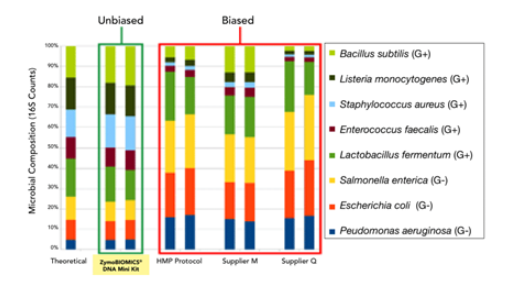
▶ Find Your Bias & Eliminate It
ZymoBIOMICS Microbial Community Standard was used to compare different DNA extraction protocols. DNA samples were profiled by 16S rRNA gene targeted sequencing.